生物信息学
20230907_从GO与KEGG富集分析中提取基因名称的方法介绍
517

在进行生物信息学分析时,基因功能富集分析是一种关键步骤,帮助研究者识别基因组数据中与特定生物学功能或通路相关的基因集合。使用如GO (Gene Ontology) 和KEGG (Kyoto Encyclopedia of Genes and Genomes) 等数据库进行富集分析后,通常会得到一系列与研究条件相关的生物通路。然而,富集结果通常提供的是通路名称和统计数据,而对于进一步的分析和实验验证,我们需要从这些富集的通路中提取出具体的基因名称。在本文中,我们将介绍如何简单而有效地从GO和KEGG富集分析结果中提取基因名称的方法。
(1) 基因功能富集分析
#install.packages("readr")
#if (!requireNamespace("BiocManager", quietly = TRUE))
#install.packages("BiocManager")
#BiocManager::install("magrittr")
library(magrittr)
#BiocManager::install("clusterProfiler")
library(clusterProfiler)
#BiocManager::install("tidyverse")
library(tidyverse)
library(readr)
library(patchwork)
library(cowplot)
library(ggplot2)
#BiocManager::install("org.Hs.eg.db")
library(org.Hs.eg.db)
#BiocManager::install(c("enrichplot", "GOplot"),ask = F,update = F)
library(enrichplot)
library(GOplot)
#install.packages("yulab.utils")
gene_list <- read.table('covid_2.go', header = F)
head(gene_list)
# 基因ID转换
id_convert <- bitr(gene_list$V1,
fromType="ENSEMBL",
toType=c("ENTREZID","ENSEMBL",'SYMBOL'),
OrgDb="org.Hs.eg.db")
head(id_convert)
# GO富集分析
go_enrich <- enrichGO(id_convert$ENTREZID,
OrgDb=org.Hs.eg.db,
ont='ALL',
pAdjustMethod='BH',
pvalueCutoff=0.05,
qvalueCutoff=0.05,
keyType='ENTREZID')
head(go_enrich@result)
# GO结果可视化 MM BP CF出现在一张图中
p1 <- dotplot(go_enrich, showCategory=3, split="ONTOLOGY")
p1
# GO结果可视化 MM BP CF在三张图中展示
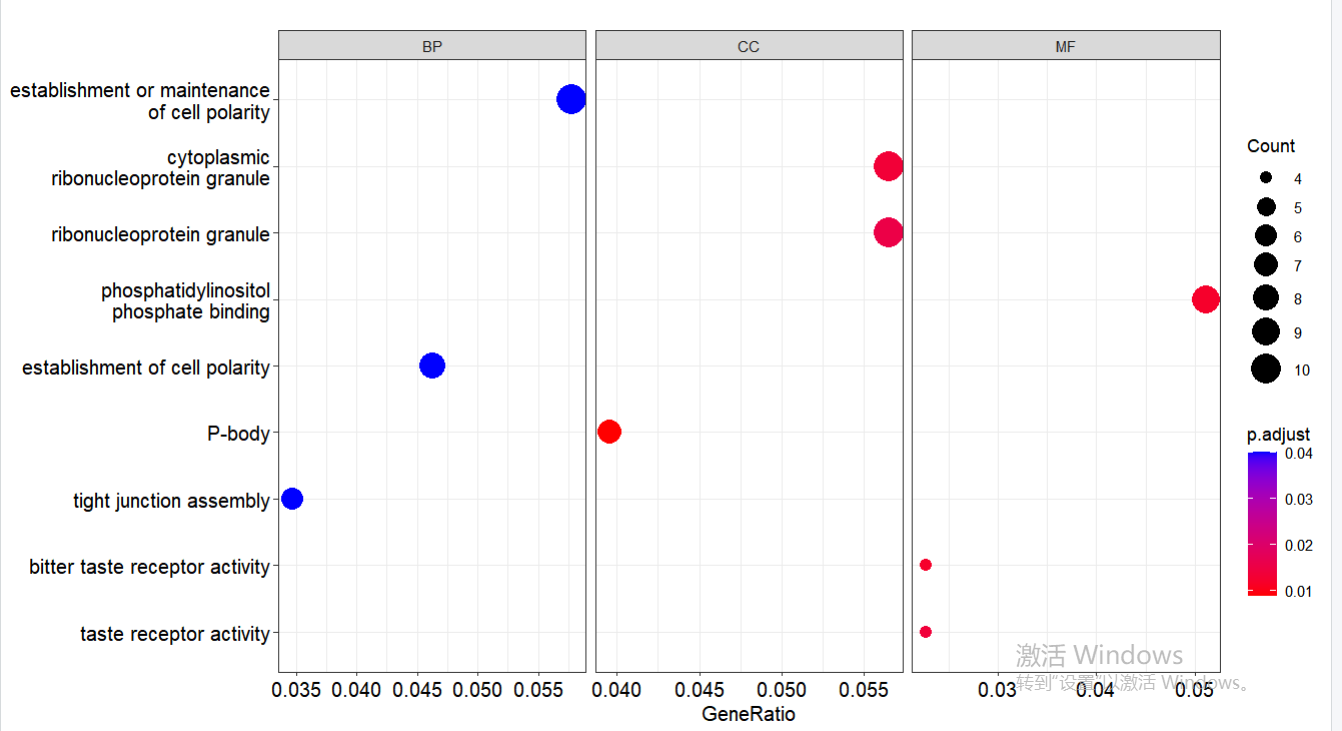
p2 <- dotplot(go_enrich, showCategory = 3, split="ONTOLOGY") +
facet_grid(.~ONTOLOGY, scale="free")
p2
ggsave("breast_cancer_ribo_go_dotplot.pdf", p1, width=15, height=5, dpi=300)
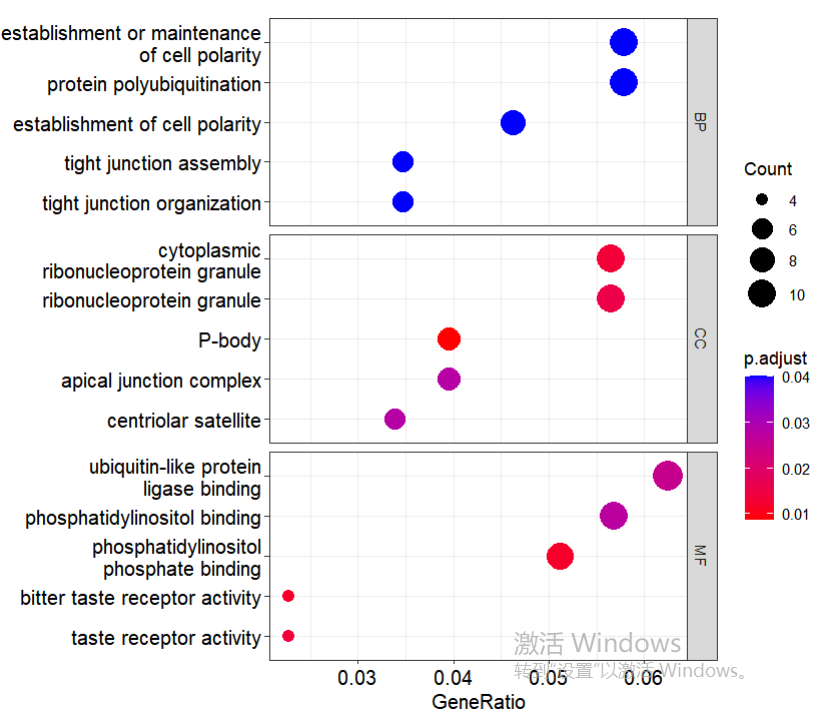
p3 <- dotplot(go_enrich, showCategory = 5, split="ONTOLOGY") +
facet_grid(ONTOLOGY~., scale="free")
p3
#############################################
# KEGG富集分析
kegg_enrich <- enrichKEGG(id_convert$ENTREZID,
organism='hsa',
pvalueCutoff=0.05,
qvalueCutoff=0.05)
head(kegg_enrich@result)
# KEGG结果可视化
p3 <- barplot(kegg_enrich, showCategory=10)
p3
p4 <- dotplot(kegg_enrich, showCategory=10)
p4
#ggsave("covid-2_dec_kegg.pdf", p4, width=7, height=7, dpi=300)> #install.packages("readr")
> #if (!requireNamespace("BiocManager", quietly = TRUE))
> #install.packages("BiocManager")
> #BiocManager::install("magrittr")
> library(magrittr)
> #BiocManager::install("clusterProfiler")
> library(clusterProfiler)
> #BiocManager::install("tidyverse")
> library(tidyverse)
> library(readr)
> library(patchwork)
> library(cowplot)
> library(ggplot2)
> #BiocManager::install("org.Hs.eg.db")
> library(org.Hs.eg.db)
> #BiocManager::install(c("enrichplot", "GOplot"),ask = F,update = F)
> library(enrichplot)
> library(GOplot)
> gene_list <- read.table('covid_2.go', header = F)
> head(gene_list)
V1
1 ENSG00000269737
2 ENSG00000215914
3 ENSG00000189339
4 ENSG00000227775
5 ENSG00000215790
6 ENSG00000008128
> # 基因ID转换
> id_convert <- bitr(gene_list$V1,
+ fromType="ENSEMBL",
+ toType=c("ENTREZID","ENSEMBL",'SYMBOL'),
+ OrgDb="org.Hs.eg.db")
'select()' returned 1:many mapping between keys and columns
Warning message:
In bitr(gene_list$V1, fromType = "ENSEMBL", toType = c("ENTREZID", :
15.97% of input gene IDs are fail to map...
> head(id_convert)
ENSEMBL ENTREZID SYMBOL
2 ENSG00000215914 8511 MMP23A
3 ENSG00000189339 728661 SLC35E2B
6 ENSG00000008128 728642 CDK11A
8 ENSG00000135250 6733 SRPK2
9 ENSG00000130856 7776 ZNF236
10 ENSG00000155974 23426 GRIP1
> # GO富集分析
> go_enrich <- enrichGO(id_convert$ENTREZID,
+ OrgDb=org.Hs.eg.db,
+ ont='ALL',
+ pAdjustMethod='BH',
+ pvalueCutoff=0.05,
+ qvalueCutoff=0.05,
+ keyType='ENTREZID')
> head(go_enrich@result)
ONTOLOGY ID Description GeneRatio BgRatio pvalue p.adjust qvalue geneID Count
GO:0007163 BP GO:0007163 establishment or maintenance of cell polarity 10/173 234/18903 6.093814e-05 0.04006962 0.03831317 2932/81609/1739/56288/5747/51057/5584/10207/55914/10806 10
GO:0120192 BP GO:0120192 tight junction assembly 6/173 75/18903 6.433027e-05 0.04006962 0.03831317 6904/1739/261734/56288/10207/79977 6
GO:0030010 BP GO:0030010 establishment of cell polarity 8/173 150/18903 7.304312e-05 0.04006962 0.03831317 2932/81609/56288/5747/51057/5584/10207/10806 8
GO:0000209 BP GO:0000209 protein polyubiquitination 10/173 246/18903 9.236316e-05 0.04006962 0.03831317 996/7326/54778/8312/57531/83737/64795/92912/7322/9520 10
GO:0120193 BP GO:0120193 tight junction organization 6/173 81/18903 9.909961e-05 0.04006962 0.03831317 6904/1739/261734/56288/10207/79977 6
GO:0045197 BP GO:0045197 establishment or maintenance of epithelial cell apical/basal polarity 5/173 51/18903 1.012534e-04 0.04006962 0.03831317 1739/56288/5584/10207/55914 5
> # GO结果可视化 MM BP CF出现在一张图中
> p1 <- dotplot(go_enrich, showCategory=3, split="ONTOLOGY")
> p1
> # GO结果可视化 MM BP CF在三张图中展示
> p2 <- dotplot(go_enrich, showCategory = 3, split="ONTOLOGY") +
+ facet_grid(.~ONTOLOGY, scale="free")
> p2
> ggsave("breast_cancer_ribo_go_dotplot.pdf", p1, width=15, height=5, dpi=300)
> p3 <- dotplot(go_enrich, showCategory = 5, split="ONTOLOGY") +
+ facet_grid(ONTOLOGY~., scale="free")
> p3
> # KEGG富集分析
> kegg_enrich <- enrichKEGG(id_convert$ENTREZID,
+ organism='hsa',
+ pvalueCutoff=0.05,
+ qvalueCutoff=0.05)
> head(kegg_enrich@result)
ID Description GeneRatio BgRatio pvalue p.adjust qvalue geneID Count
hsa04742 hsa04742 Taste transduction 6/78 86/8586 0.0001221859 0.01991631 0.01954975 259296/259295/259294/259290/50840/5137 6
hsa04120 hsa04120 Ubiquitin mediated proteolysis 6/78 142/8586 0.0017797275 0.14504779 0.14237820 996/26091/7326/83737/92912/7322 6
hsa04390 hsa04390 Hippo signaling pathway 6/78 157/8586 0.0029509254 0.16033362 0.15738269 2932/1739/8312/56288/5584/10207 6
hsa04141 hsa04141 Protein processing in endoplasmic reticulum 6/78 170/8586 0.0043672144 0.17796399 0.17468858 11231/7326/23193/3301/29979/7322 6
hsa04360 hsa04360 Axon guidance 6/78 182/8586 0.0060725451 0.19796497 0.19432144 2932/56288/2771/5747/10509/2050 6
hsa04144 hsa04144 Endocytosis 7/78 250/8586 0.0074292397 0.20182768 0.19811306 56288/83737/116984/30011/5584/55680/58533 7
> # KEGG结果可视化
> p3 <- barplot(kegg_enrich, showCategory=10)
> p3
> p4 <- dotplot(kegg_enrich, showCategory=10)
> p4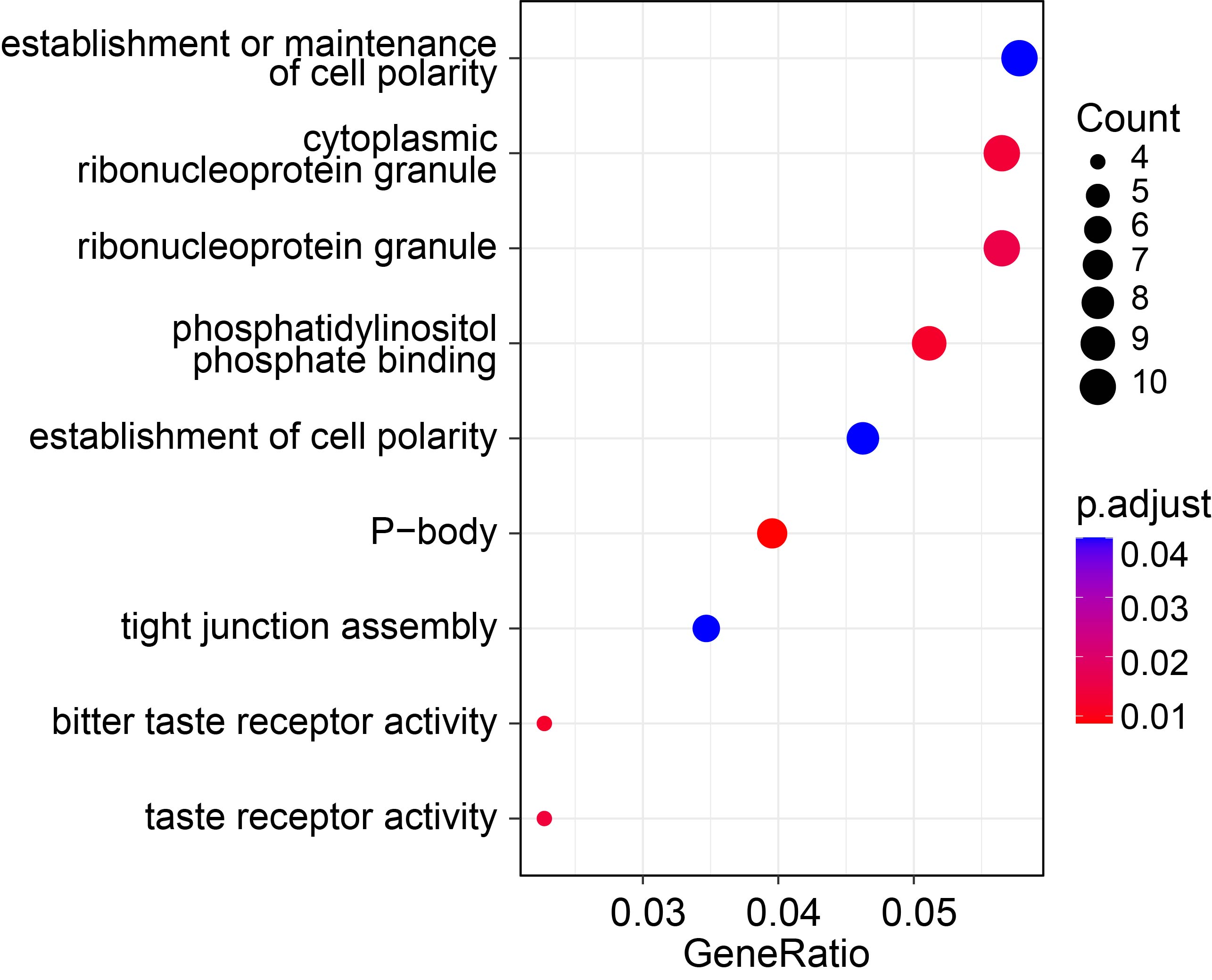
(2)提取富集到的与特定生物功能或通路相关的基因
library(dplyr)
library(AnnotationDbi)
library(org.Hs.eg.db)
head(id_convert)
head(go_enrich@result)
head(go_enrich@result$geneID)
head(go_enrich@result$Description)
# 从go_enrich结果中筛选BP前3项
bp_top3 <- go_enrich %>% filter(ONTOLOGY == "BP") %>% head(3)
# 从go_enrich结果中筛选CC前3项
cc_top3 <- go_enrich %>% filter(ONTOLOGY == "CC") %>% head(3)
# 从go_enrich结果中筛选MF前3项
mf_top3 <- go_enrich %>% filter(ONTOLOGY == "MF") %>% head(3)
# 组合新的结果
new_result <- bind_rows(bp_top3, cc_top3, mf_top3)
# 查看新结果
head(new_result)
#head(new_result)
#head(new_result$geneID)
# 提取geneID
geneids <- new_result$geneID
head(geneids)
#####################################################
# 在GO富集的term后面加上该通路的每个基因的SYMBOL基因名
#####################################################
# 对每个geneID进行分割并映射到symbol或者SYMBOL
mapped_ids <- lapply(geneids, function(x) {
genes <- unlist(strsplit(x, "/"))
#mapped <- bitr(genes, fromType="ENSEMBL", toType="SYMBOL", OrgDb="org.Hs.eg.db")
#mapped <- bitr(genes, fromType="ENTREZID", toType="SYMBOL", OrgDb="org.Hs.eg.db")
#return(mapped$SYMBOL)
mapped <- bitr(genes, fromType="ENTREZID", toType="SYMBOL", OrgDb="org.Hs.eg.db")
return(mapped$SYMBOL)
})
head(mapped_ids)
# 拼接GENERATED基因符号
new_result$geneID <- sapply(mapped_ids, function(x) paste(x, collapse=","))
# 查看结果
head(new_result)
head(new_result[,c(1,2,3,9)])
#####################################################
# 在GO富集的term后面加上该通路的每个基因的ENSEMBL编号
#####################################################
# 对每个geneID进行分割并映射到symbol或者SYMBOL
mapped_ids <- lapply(geneids, function(x) {
genes <- unlist(strsplit(x, "/"))
#mapped <- bitr(genes, fromType="ENSEMBL", toType="SYMBOL", OrgDb="org.Hs.eg.db")
#mapped <- bitr(genes, fromType="ENTREZID", toType="SYMBOL", OrgDb="org.Hs.eg.db")
#return(mapped$SYMBOL)
mapped <- bitr(genes, fromType="ENTREZID", toType="ENSEMBL", OrgDb="org.Hs.eg.db")
return(mapped$ENSEMBL)
})
head(mapped_ids)
# 拼接GENERATED基因符号
new_result$geneID <- sapply(mapped_ids, function(x) paste(x, collapse=","))
# 查看结果
head(new_result)
head(new_result$geneID)
head(new_result[,c(1,2,3,9)])
#####################################################
# 将new_result表格中的 geneID切割成单个基因,并且保存到文件中。这些保存的基因就是参与GO富集分析pdf图中所有通路的基因
#####################################################
# 提取基因IDs
gene_ids <- new_result$geneID
# 初始化记录每个基因的向量
genes <- vector("character", 0)
# 遍历每个基因向量
for(i in seq_along(gene_ids)){
# 拆分基因ID
id_vector <- strsplit(gene_ids[[i]], ",")[[1]]
# 按行追加到genes向量
genes <- c(genes, id_vector)
}
# 去重
genes <- unique(genes)
head(genes)
# 写入文件
writeLines(genes, con="genes.txt")
(3)代码执行结果
> library(dplyr)
> library(AnnotationDbi)
> library(org.Hs.eg.db)
> head(id_convert)
ENSEMBL ENTREZID SYMBOL
2 ENSG00000215914 8511 MMP23A
3 ENSG00000189339 728661 SLC35E2B
6 ENSG00000008128 728642 CDK11A
8 ENSG00000135250 6733 SRPK2
9 ENSG00000130856 7776 ZNF236
10 ENSG00000155974 23426 GRIP1
> head(go_enrich@result)
ONTOLOGY ID Description GeneRatio BgRatio pvalue p.adjust qvalue geneID Count
GO:0007163 BP GO:0007163 establishment or maintenance of cell polarity 10/173 234/18903 6.093814e-05 0.04006962 0.03831317 2932/81609/1739/56288/5747/51057/5584/10207/55914/10806 10
GO:0120192 BP GO:0120192 tight junction assembly 6/173 75/18903 6.433027e-05 0.04006962 0.03831317 6904/1739/261734/56288/10207/79977 6
GO:0030010 BP GO:0030010 establishment of cell polarity 8/173 150/18903 7.304312e-05 0.04006962 0.03831317 2932/81609/56288/5747/51057/5584/10207/10806 8
GO:0000209 BP GO:0000209 protein polyubiquitination 10/173 246/18903 9.236316e-05 0.04006962 0.03831317 996/7326/54778/8312/57531/83737/64795/92912/7322/9520 10
GO:0120193 BP GO:0120193 tight junction organization 6/173 81/18903 9.909961e-05 0.04006962 0.03831317 6904/1739/261734/56288/10207/79977 6
GO:0045197 BP GO:0045197 establishment or maintenance of epithelial cell apical/basal polarity 5/173 51/18903 1.012534e-04 0.04006962 0.03831317 1739/56288/5584/10207/55914 5
> head(go_enrich@result$geneID)
[1] "2932/81609/1739/56288/5747/51057/5584/10207/55914/10806" "6904/1739/261734/56288/10207/79977" "2932/81609/56288/5747/51057/5584/10207/10806" "996/7326/54778/8312/57531/83737/64795/92912/7322/9520"
[5] "6904/1739/261734/56288/10207/79977" "1739/56288/5584/10207/55914"
> head(go_enrich@result$Description)
[1] "establishment or maintenance of cell polarity" "tight junction assembly" "establishment of cell polarity"
[4] "protein polyubiquitination" "tight junction organization" "establishment or maintenance of epithelial cell apical/basal polarity"
> # 从go_enrich结果中筛选BP前3项
> bp_top3 <- go_enrich %>% filter(ONTOLOGY == "BP") %>% head(3)
> # 从go_enrich结果中筛选CC前3项
> cc_top3 <- go_enrich %>% filter(ONTOLOGY == "CC") %>% head(3)
> # 从go_enrich结果中筛选MF前3项
> mf_top3 <- go_enrich %>% filter(ONTOLOGY == "MF") %>% head(3)
> # 组合新的结果
> new_result <- bind_rows(bp_top3, cc_top3, mf_top3)
> # 查看新结果
> head(new_result)
ONTOLOGY ID Description GeneRatio BgRatio pvalue p.adjust qvalue geneID Count
GO:0007163 BP GO:0007163 establishment or maintenance of cell polarity 10/173 234/18903 6.093814e-05 0.040069621 0.038313175 2932/81609/1739/56288/5747/51057/5584/10207/55914/10806 10
GO:0120192 BP GO:0120192 tight junction assembly 6/173 75/18903 6.433027e-05 0.040069621 0.038313175 6904/1739/261734/56288/10207/79977 6
GO:0030010 BP GO:0030010 establishment of cell polarity 8/173 150/18903 7.304312e-05 0.040069621 0.038313175 2932/81609/56288/5747/51057/5584/10207/10806 8
GO:0000932 CC GO:0000932 P-body 7/177 97/19869 2.587477e-05 0.008745672 0.007653484 55833/27161/255967/10643/23034/64506/11030 7
GO:0036464 CC GO:0036464 cytoplasmic ribonucleoprotein granule 10/177 248/19869 7.953890e-05 0.013442074 0.011763385 55833/9960/27161/255967/10643/23034/64506/11030/5936/6311 10
GO:0035770 CC GO:0035770 ribonucleoprotein granule 10/177 265/19869 1.372325e-04 0.015461524 0.013530640 55833/9960/27161/255967/10643/23034/64506/11030/5936/6311 10
> # 提取geneID
> geneids <- new_result$geneID
> head(geneids)
[1] "2932/81609/1739/56288/5747/51057/5584/10207/55914/10806" "6904/1739/261734/56288/10207/79977" "2932/81609/56288/5747/51057/5584/10207/10806" "55833/27161/255967/10643/23034/64506/11030"
[5] "55833/9960/27161/255967/10643/23034/64506/11030/5936/6311" "55833/9960/27161/255967/10643/23034/64506/11030/5936/6311"
#####################################################
# 在GO富集的term后面加上该通路的每个基因的SYMBOL基因名
#####################################################
> # 对每个geneID进行分割并映射到symbol或者SYMBOL
> mapped_ids <- lapply(geneids, function(x) {
+ genes <- unlist(strsplit(x, "/"))
+ #mapped <- bitr(genes, fromType="ENSEMBL", toType="SYMBOL", OrgDb="org.Hs.eg.db")
+ #mapped <- bitr(genes, fromType="ENTREZID", toType="SYMBOL", OrgDb="org.Hs.eg.db")
+ #return(mapped$SYMBOL)
+ mapped <- bitr(genes, fromType="ENTREZID", toType="SYMBOL", OrgDb="org.Hs.eg.db")
+ return(mapped$SYMBOL)
+ })
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
> head(mapped_ids)
[[1]]
[1] "GSK3B" "SNX27" "DLG1" "PARD3" "PTK2" "WDPCP" "PRKCI" "PATJ" "ERBIN" "SDCCAG8"
[[2]]
[1] "TBCD" "DLG1" "NPHP4" "PARD3" "PATJ" "GRHL2"
[[3]]
[1] "GSK3B" "SNX27" "PARD3" "PTK2" "WDPCP" "PRKCI" "PATJ" "SDCCAG8"
[[4]]
[1] "UBAP2" "AGO2" "PAN3" "IGF2BP3" "SAMD4A" "CPEB1" "RBPMS"
[[5]]
[1] "UBAP2" "USP3" "AGO2" "PAN3" "IGF2BP3" "SAMD4A" "CPEB1" "RBPMS" "RBM4" "ATXN2"
[[6]]
[1] "UBAP2" "USP3" "AGO2" "PAN3" "IGF2BP3" "SAMD4A" "CPEB1" "RBPMS" "RBM4" "ATXN2"
> # 拼接GENERATED基因符号
> new_result$geneID <- sapply(mapped_ids, function(x) paste(x, collapse=","))
> # 查看结果
> head(new_result)
ONTOLOGY ID Description GeneRatio BgRatio pvalue p.adjust qvalue geneID Count
GO:0007163 BP GO:0007163 establishment or maintenance of cell polarity 10/173 234/18903 6.093814e-05 0.040069621 0.038313175 GSK3B,SNX27,DLG1,PARD3,PTK2,WDPCP,PRKCI,PATJ,ERBIN,SDCCAG8 10
GO:0120192 BP GO:0120192 tight junction assembly 6/173 75/18903 6.433027e-05 0.040069621 0.038313175 TBCD,DLG1,NPHP4,PARD3,PATJ,GRHL2 6
GO:0030010 BP GO:0030010 establishment of cell polarity 8/173 150/18903 7.304312e-05 0.040069621 0.038313175 GSK3B,SNX27,PARD3,PTK2,WDPCP,PRKCI,PATJ,SDCCAG8 8
GO:0000932 CC GO:0000932 P-body 7/177 97/19869 2.587477e-05 0.008745672 0.007653484 UBAP2,AGO2,PAN3,IGF2BP3,SAMD4A,CPEB1,RBPMS 7
GO:0036464 CC GO:0036464 cytoplasmic ribonucleoprotein granule 10/177 248/19869 7.953890e-05 0.013442074 0.011763385 UBAP2,USP3,AGO2,PAN3,IGF2BP3,SAMD4A,CPEB1,RBPMS,RBM4,ATXN2 10
GO:0035770 CC GO:0035770 ribonucleoprotein granule 10/177 265/19869 1.372325e-04 0.015461524 0.013530640 UBAP2,USP3,AGO2,PAN3,IGF2BP3,SAMD4A,CPEB1,RBPMS,RBM4,ATXN2 10
###########这是我们想要的结果样式
> head(new_result[,c(1,2,3,9)])
ONTOLOGY ID Description geneID
GO:0007163 BP GO:0007163 establishment or maintenance of cell polarity GSK3B,SNX27,DLG1,PARD3,PTK2,WDPCP,PRKCI,PATJ,ERBIN,SDCCAG8
GO:0120192 BP GO:0120192 tight junction assembly TBCD,DLG1,NPHP4,PARD3,PATJ,GRHL2
GO:0030010 BP GO:0030010 establishment of cell polarity GSK3B,SNX27,PARD3,PTK2,WDPCP,PRKCI,PATJ,SDCCAG8
GO:0000932 CC GO:0000932 P-body UBAP2,AGO2,PAN3,IGF2BP3,SAMD4A,CPEB1,RBPMS
GO:0036464 CC GO:0036464 cytoplasmic ribonucleoprotein granule UBAP2,USP3,AGO2,PAN3,IGF2BP3,SAMD4A,CPEB1,RBPMS,RBM4,ATXN2
GO:0035770 CC GO:0035770 ribonucleoprotein granule UBAP2,USP3,AGO2,PAN3,IGF2BP3,SAMD4A,CPEB1,RBPMS,RBM4,ATXN2
#####################################################
# 在GO富集的term后面加上该通路的每个基因的ENSEMBL编号
#####################################################
> # 对每个geneID进行分割并映射到symbol或者SYMBOL
> mapped_ids <- lapply(geneids, function(x) {
+ genes <- unlist(strsplit(x, "/"))
+ #mapped <- bitr(genes, fromType="ENSEMBL", toType="SYMBOL", OrgDb="org.Hs.eg.db")
+ #mapped <- bitr(genes, fromType="ENTREZID", toType="SYMBOL", OrgDb="org.Hs.eg.db")
+ #return(mapped$SYMBOL)
+ mapped <- bitr(genes, fromType="ENTREZID", toType="ENSEMBL", OrgDb="org.Hs.eg.db")
+ return(mapped$ENSEMBL)
+ })
'select()' returned 1:many mapping between keys and columns
'select()' returned 1:many mapping between keys and columns
'select()' returned 1:many mapping between keys and columns
'select()' returned 1:many mapping between keys and columns
'select()' returned 1:many mapping between keys and columns
'select()' returned 1:many mapping between keys and columns
'select()' returned 1:many mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:many mapping between keys and columns
> head(mapped_ids)
[[1]]
[1] "ENSG00000082701" "ENSG00000143376" "ENSG00000075711" "ENSG00000148498" "ENSG00000169398" "ENSG00000143951" "ENSG00000163558" "ENSG00000132849" "ENSG00000112851" "ENSG00000054282" "ENSG00000276111"
[[2]]
[1] "ENSG00000141556" "ENSG00000278759" "ENSG00000075711" "ENSG00000131697" "ENSG00000148498" "ENSG00000132849" "ENSG00000083307"
[[3]]
[1] "ENSG00000082701" "ENSG00000143376" "ENSG00000148498" "ENSG00000169398" "ENSG00000143951" "ENSG00000163558" "ENSG00000132849" "ENSG00000054282" "ENSG00000276111"
[[4]]
[1] "ENSG00000137073" "ENSG00000123908" "ENSG00000152520" "ENSG00000136231" "ENSG00000020577" "ENSG00000214575" "ENSG00000277445" "ENSG00000157110"
[[5]]
[1] "ENSG00000137073" "ENSG00000140455" "ENSG00000123908" "ENSG00000152520" "ENSG00000136231" "ENSG00000020577" "ENSG00000214575" "ENSG00000277445" "ENSG00000157110" "ENSG00000173933" "ENSG00000204842"
[[6]]
[1] "ENSG00000137073" "ENSG00000140455" "ENSG00000123908" "ENSG00000152520" "ENSG00000136231" "ENSG00000020577" "ENSG00000214575" "ENSG00000277445" "ENSG00000157110" "ENSG00000173933" "ENSG00000204842"
> # 拼接GENERATED基因符号
> new_result$geneID <- sapply(mapped_ids, function(x) paste(x, collapse=","))
> # 查看结果
> head(new_result)
ONTOLOGY ID Description GeneRatio BgRatio pvalue p.adjust qvalue
GO:0007163 BP GO:0007163 establishment or maintenance of cell polarity 10/173 234/18903 6.093814e-05 0.040069621 0.038313175
GO:0120192 BP GO:0120192 tight junction assembly 6/173 75/18903 6.433027e-05 0.040069621 0.038313175
GO:0030010 BP GO:0030010 establishment of cell polarity 8/173 150/18903 7.304312e-05 0.040069621 0.038313175
GO:0000932 CC GO:0000932 P-body 7/177 97/19869 2.587477e-05 0.008745672 0.007653484
GO:0036464 CC GO:0036464 cytoplasmic ribonucleoprotein granule 10/177 248/19869 7.953890e-05 0.013442074 0.011763385
GO:0035770 CC GO:0035770 ribonucleoprotein granule 10/177 265/19869 1.372325e-04 0.015461524 0.013530640
geneID Count
GO:0007163 ENSG00000082701,ENSG00000143376,ENSG00000075711,ENSG00000148498,ENSG00000169398,ENSG00000143951,ENSG00000163558,ENSG00000132849,ENSG00000112851,ENSG00000054282,ENSG00000276111 10
GO:0120192 ENSG00000141556,ENSG00000278759,ENSG00000075711,ENSG00000131697,ENSG00000148498,ENSG00000132849,ENSG00000083307 6
GO:0030010 ENSG00000082701,ENSG00000143376,ENSG00000148498,ENSG00000169398,ENSG00000143951,ENSG00000163558,ENSG00000132849,ENSG00000054282,ENSG00000276111 8
GO:0000932 ENSG00000137073,ENSG00000123908,ENSG00000152520,ENSG00000136231,ENSG00000020577,ENSG00000214575,ENSG00000277445,ENSG00000157110 7
GO:0036464 ENSG00000137073,ENSG00000140455,ENSG00000123908,ENSG00000152520,ENSG00000136231,ENSG00000020577,ENSG00000214575,ENSG00000277445,ENSG00000157110,ENSG00000173933,ENSG00000204842 10
GO:0035770 ENSG00000137073,ENSG00000140455,ENSG00000123908,ENSG00000152520,ENSG00000136231,ENSG00000020577,ENSG00000214575,ENSG00000277445,ENSG00000157110,ENSG00000173933,ENSG00000204842 10
> head(new_result$geneID)
[1] "ENSG00000082701,ENSG00000143376,ENSG00000075711,ENSG00000148498,ENSG00000169398,ENSG00000143951,ENSG00000163558,ENSG00000132849,ENSG00000112851,ENSG00000054282,ENSG00000276111"
[2] "ENSG00000141556,ENSG00000278759,ENSG00000075711,ENSG00000131697,ENSG00000148498,ENSG00000132849,ENSG00000083307"
[3] "ENSG00000082701,ENSG00000143376,ENSG00000148498,ENSG00000169398,ENSG00000143951,ENSG00000163558,ENSG00000132849,ENSG00000054282,ENSG00000276111"
[4] "ENSG00000137073,ENSG00000123908,ENSG00000152520,ENSG00000136231,ENSG00000020577,ENSG00000214575,ENSG00000277445,ENSG00000157110"
[5] "ENSG00000137073,ENSG00000140455,ENSG00000123908,ENSG00000152520,ENSG00000136231,ENSG00000020577,ENSG00000214575,ENSG00000277445,ENSG00000157110,ENSG00000173933,ENSG00000204842"
[6] "ENSG00000137073,ENSG00000140455,ENSG00000123908,ENSG00000152520,ENSG00000136231,ENSG00000020577,ENSG00000214575,ENSG00000277445,ENSG00000157110,ENSG00000173933,ENSG00000204842"
###########这是我们想要的结果样式
> head(new_result[,c(1,2,3,9)])
ONTOLOGY ID Description geneID
GO:0007163 BP GO:0007163 establishment or maintenance of cell polarity ENSG00000082701,ENSG00000143376,ENSG00000075711,ENSG00000148498,ENSG00000169398,ENSG00000143951,ENSG00000163558,ENSG00000132849,ENSG00000112851,ENSG00000054282,ENSG00000276111
GO:0120192 BP GO:0120192 tight junction assembly ENSG00000141556,ENSG00000278759,ENSG00000075711,ENSG00000131697,ENSG00000148498,ENSG00000132849,ENSG00000083307
GO:0030010 BP GO:0030010 establishment of cell polarity ENSG00000082701,ENSG00000143376,ENSG00000148498,ENSG00000169398,ENSG00000143951,ENSG00000163558,ENSG00000132849,ENSG00000054282,ENSG00000276111
GO:0000932 CC GO:0000932 P-body ENSG00000137073,ENSG00000123908,ENSG00000152520,ENSG00000136231,ENSG00000020577,ENSG00000214575,ENSG00000277445,ENSG00000157110
GO:0036464 CC GO:0036464 cytoplasmic ribonucleoprotein granule ENSG00000137073,ENSG00000140455,ENSG00000123908,ENSG00000152520,ENSG00000136231,ENSG00000020577,ENSG00000214575,ENSG00000277445,ENSG00000157110,ENSG00000173933,ENSG00000204842
GO:0035770 CC GO:0035770 ribonucleoprotein granule ENSG00000137073,ENSG00000140455,ENSG00000123908,ENSG00000152520,ENSG00000136231,ENSG00000020577,ENSG00000214575,ENSG00000277445,ENSG00000157110,ENSG00000173933,ENSG00000204842
#####################################################
# 将new_result表格中的 geneID切割成单个基因,并且保存到文件中。这些保存的基因就是参与GO富集分析pdf图中所有通路的基因
#####################################################
> # 提取基因IDs
> gene_ids <- new_result$geneID
> head(gene_ids)
[1] "ENSG00000082701,ENSG00000143376,ENSG00000075711,ENSG00000148498,ENSG00000169398,ENSG00000143951,ENSG00000163558,ENSG00000132849,ENSG00000112851,ENSG00000054282,ENSG00000276111"
[2] "ENSG00000141556,ENSG00000278759,ENSG00000075711,ENSG00000131697,ENSG00000148498,ENSG00000132849,ENSG00000083307"
[3] "ENSG00000082701,ENSG00000143376,ENSG00000148498,ENSG00000169398,ENSG00000143951,ENSG00000163558,ENSG00000132849,ENSG00000054282,ENSG00000276111"
[4] "ENSG00000137073,ENSG00000123908,ENSG00000152520,ENSG00000136231,ENSG00000020577,ENSG00000214575,ENSG00000277445,ENSG00000157110"
[5] "ENSG00000137073,ENSG00000140455,ENSG00000123908,ENSG00000152520,ENSG00000136231,ENSG00000020577,ENSG00000214575,ENSG00000277445,ENSG00000157110,ENSG00000173933,ENSG00000204842"
[6] "ENSG00000137073,ENSG00000140455,ENSG00000123908,ENSG00000152520,ENSG00000136231,ENSG00000020577,ENSG00000214575,ENSG00000277445,ENSG00000157110,ENSG00000173933,ENSG00000204842"
> # 初始化记录每个基因的向量
> genes <- vector("character", 0)
> # 遍历每个基因向量
> for(i in seq_along(gene_ids)){
+ # 拆分基因ID
+ id_vector <- strsplit(gene_ids[[i]], ",")[[1]]
+ # 按行追加到genes向量
+ genes <- c(genes, id_vector)
+ }
> # 去重
> genes <- unique(genes)
> head(genes)
[1] "ENSG00000082701" "ENSG00000143376" "ENSG00000075711" "ENSG00000148498" "ENSG00000169398" "ENSG00000143951"> # 写入文件
> writeLines(genes, con="genes.txt")
标签:
bioinformatics
北京 天气
晴
0℃


