R语言编程
20231005_桑基气泡图:深度解读GO富集分析的五维度信息
1465

在生物信息学领域,基因本体论(Gene Ontology,GO)富集分析是一个常用的方法,用于解析基因集合的功能特性。然而,传统的GO富集分析结果可视化方法往往只能展示有限的信息,并未充分利用富集分析的多维度信息。为了更好地展示和理解这些多维度信息,我们选择使用桑基气泡图。
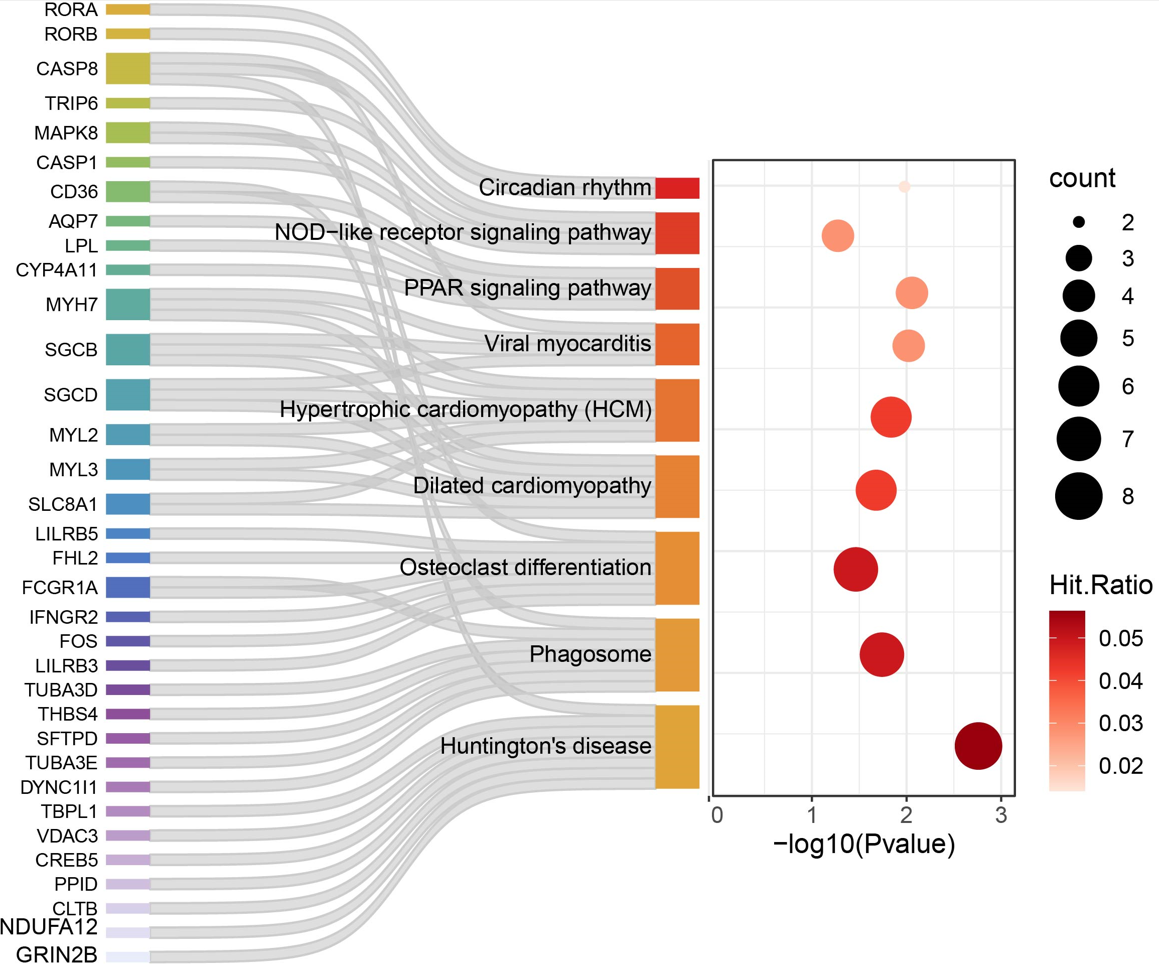
桑基气泡图是一种新型的数据可视化工具,它结合了桑基图(Sankey Diagram)和气泡图(Bubble Chart)的特性,能够展示更多维度的信息。桑基气泡图不仅可以展示基因在不同路径中的分布、数量和显著性,还可以通过颜色和大小的变化直观地展示统计显著性和基因数量的变化。更重要的是,桑基气泡图可以展示基因ID的流动,帮助我们理解基因在不同生物过程中的角色,以及基因之间的关系。
本文将详细介绍桑基气泡图的原理和作图方法,并以实例展示如何使用桑基气泡图进行GO富集分析的结果可视化,以期通过更直观、更深入的方式理解基因的功能特性。
软件包安装:
# 以管理员的身份运行Rstudio
#R 4.3.1
#Rtools 4.3
#Biocmanager 3.17
#BiocManager::install("tidyverse")
library(tidyverse)
#BiocManager::install("ggsankey")
#install.packages("ggsankey")
library(devtools)
#devtools::install_github("davidsjoberg/ggsankey")
library(ggsankey)
library(ggplot2)
#BiocManager::install("cols4all")
library(cols4all)
#BiocManager::install("cowplot")
library(cowplot)
#install.packages(c("shinyjs", "kableExtra", "colorblindcheck"))
library(shinyjs)
library(kableExtra)
library(colorblindcheck)
测试数据:
> kegg <- read.csv('kegg_result.csv',header = T)
> kegg
pathNames count Pvalue Hit.Ratio
1 Circadian rhythm 2 0.010497623 0.01408451
2 NOD-like receptor signaling pathway 4 0.053032550 0.02816901
3 PPAR signaling pathway 4 0.008761306 0.02816901
4 Viral myocarditis 4 0.009484362 0.02816901
5 Hypertrophic cardiomyopathy (HCM) 6 0.014532337 0.04225352
6 Dilated cardiomyopathy 6 0.020924238 0.04225352
7 Osteoclast differentiation 7 0.034284302 0.04929578
8 Phagosome 7 0.018180176 0.04929578
9 Huntington's disease 8 0.001736229 0.05633803
> sankey <- read.csv('sankey_result.csv',header = T)
> sankey
metamolites pathNames
1 RORA Circadian rhythm
2 RORB Circadian rhythm
3 CASP8 NOD-like receptor signaling pathway
4 TRIP6 NOD-like receptor signaling pathway
5 MAPK8 NOD-like receptor signaling pathway
6 CASP1 NOD-like receptor signaling pathway
7 CD36 PPAR signaling pathway
8 AQP7 PPAR signaling pathway
9 LPL PPAR signaling pathway
10 CYP4A11 PPAR signaling pathway
11 CASP8 Viral myocarditis
12 MYH7 Viral myocarditis
13 SGCB Viral myocarditis
14 SGCD Viral myocarditis
15 MYH7 Hypertrophic cardiomyopathy (HCM)
16 MYL2 Hypertrophic cardiomyopathy (HCM)
17 MYL3 Hypertrophic cardiomyopathy (HCM)
18 SGCB Hypertrophic cardiomyopathy (HCM)
19 SGCD Hypertrophic cardiomyopathy (HCM)
20 SLC8A1 Hypertrophic cardiomyopathy (HCM)
21 MYH7 Dilated cardiomyopathy
22 MYL2 Dilated cardiomyopathy
23 MYL3 Dilated cardiomyopathy
24 SGCB Dilated cardiomyopathy
25 SGCD Dilated cardiomyopathy
26 SLC8A1 Dilated cardiomyopathy
27 LILRB5 Osteoclast differentiation
28 MAPK8 Osteoclast differentiation
29 FHL2 Osteoclast differentiation
30 FCGR1A Osteoclast differentiation
31 IFNGR2 Osteoclast differentiation
32 FOS Osteoclast differentiation
33 LILRB3 Osteoclast differentiation
34 TUBA3D Phagosome
35 THBS4 Phagosome
36 SFTPD Phagosome
37 CD36 Phagosome
38 FCGR1A Phagosome
39 TUBA3E Phagosome
40 DYNC1I1 Phagosome
41 TBPL1 Huntington's disease
42 CASP8 Huntington's disease
43 VDAC3 Huntington's disease
44 CREB5 Huntington's disease
45 PPID Huntington's disease
46 CLTB Huntington's disease
47 NDUFA12 Huntington's disease
48 GRIN2B Huntington's disease代码运行:
kegg
kegg$pathNames <- factor(kegg$pathNames,levels = rev(kegg$pathNames))
#基础富集气泡图:
p1 <- ggplot() +
geom_point(data = kegg,
aes(x = -log10(Pvalue),
y = pathNames,
size = count,
color = Hit.Ratio)) + # 气泡大小及颜色设置
theme_bw() +
labs(x = "-log10(Pvalue)",
y = "")
p1
dim(kegg)
#数据处理:
kegg2 <- kegg[9:1,] #先调转数据框方向
kegg2 <- kegg2 %>%
mutate(ymax = cumsum(count)) %>% #ymax为Width列的累加和
mutate(ymin = ymax -count) %>%
mutate(label = (ymin + ymax)/2) #取xmin和xmax的中心位置作为标签位置
head(kegg2)
p2 <- ggplot() +
geom_point(data = kegg2,
aes(x = -log10(Pvalue),
y = label, #替换y轴列为数值
size = count,
color = Hit.Ratio)) +
theme_bw() +
labs(x = "-log10(Pvalue)",
y = "")
p2
#自定义主题与配色修改:
mytheme <- theme(axis.title = element_text(size = 13),
axis.text = element_text(size = 11),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
legend.title = element_text(size = 13),
legend.text = element_text(size = 11))
p3 <- ggplot() +
geom_point(data = kegg2,
aes(x = -log10(Pvalue),
y = label,
size = count,
color = Hit.Ratio)) +
scale_size_continuous(range=c(2,10)) + #调整气泡大小范围(默认尺寸部分过小)
scale_y_continuous(expand = c(0,0.1),limits = c(0,52)) +
#scale_x_continuous(limits = c(0.57,2.5)) +
scale_x_continuous(limits = c(0.1,3)) +
scale_colour_distiller(palette = "Reds", direction = 1) + #更改配色
labs(x = "-log10(Pvalue)",
y = "") +
theme_bw() +
mytheme
p3
#将数据转换为绘图所需格式:
head(sankey)
df <- sankey %>%
make_long(metamolites, pathNames)
head(df)
#指定绘图顺序(转换为因子):
df$node <- factor(df$node,levels = c(sankey$pathNames %>% unique()%>% rev(),
sankey$metamolites %>% unique() %>% rev()))
df
#自定义配色:
c4a_gui()
head(sankey)
dim(sankey)
length(df$node)
length(mycol)
length(unique(df$node))
#下面的43与length(unique(df$node))数值相同,此处尤其重要,不能写错
mycol <- c4a('rainbow_wh_rd',43)
#mycol
#绘图:
p4 <- ggplot(df, aes(x = x,
next_x = next_x,
node = node,
next_node = next_node,
fill = node,
label = node)) +
geom_sankey(flow.alpha = 0.5,
flow.fill = 'grey',
flow.color = 'grey80', #条带描边色
node.fill = mycol, #节点填充色
smooth = 8,
width = 0.08) +
geom_sankey_text(size = 3.2,
color = "black")+
theme_void() +
theme(legend.position = 'none')
p4
p44 <- ggplot(df, aes(x = x,
next_x = next_x,
node = node,
next_node = next_node,
fill = node,
label = node)) +
geom_sankey(flow.alpha = 0.5,
flow.fill = 'grey',
flow.color = 'grey80', #条带描边色
node.fill = mycol, #节点填充色
smooth = 8,
width = 0.1) +
geom_sankey_text(size = 3.2,
color = "black")+
theme_void() +
theme(legend.position = 'none')
p44
#通过调整页边距在桑基图右侧先留出空白:
p5 <- p4 + theme(plot.margin = unit(c(0,5,0,0),units="cm"))
p5 <- p4 + theme(plot.margin = unit(c(0,5,0,0),units="cm"))
p5
#拼图(在p5的空白位置中插入p3):
#ggdraw() + draw_plot(p5) + draw_plot(p3, scale = 0.5, x = 0.62, y=-0.21, width=0.48, height=1.37)
p6 <- ggdraw() + draw_plot(p5) + draw_plot(p3, scale = 0.5, x = 0.50, y=-0.21, width=0.68, height=1.37)
p6
ggsave("sankey_dot_plot.pdf", p6, width = 10, height = 7, dpi=300)
标签:
rstudio
北京 天气
晴
-7℃


