R语言编程
20230705_gggenes包:ggplot2下的基因与蛋白质结构可视化工具

在生物信息学和分子生物学研究中,对基因和蛋白质结构的可视化分析有助于解析它们的功能、进化关系以及相互作用。然而,传统的绘图工具可能难以满足研究人员日益增长的数据处理和分析需求。在本文中,我们将介绍一个名为gggenes的R包,它基于流行的ggplot2库,帮助研究者轻松绘制基因和蛋白质结构。我们将详细介绍gggenes包的安装与使用方法。
(01) gggenes软件包安装:
devtools::install_github("wilkox/gggenes")
or
BiocManager::install("gggenes")(02) gggenes软件包测试数据:
molecule列表示基因所属的基因组名称;gene列展示了基因的名称;start列显示了基因在基因组中的起始位置,需要注意的是负链上的基因起始位置表示方法与BED文件有所不同;end列揭示了基因在基因组中的结束位置,对于负链上的基因,起始位置的绝对值会大于结束位置;而strand列则表示基因所在的链,即正链或负链,这一列为可选项。
> head(example_genes)
molecule gene start end strand orientation
1 Genome1 genA 15389 17299 reverse 1
2 Genome1 genB 17301 18161 forward 0
3 Genome1 genC 18176 18640 reverse 1
4 Genome1 genD 18641 18985 forward 0
5 Genome1 genE 18999 20078 reverse 1
6 Genome1 genF 20086 20451 forward 1
7 Genome1 protC 22777 22989 forward 1
8 Genome1 protD 22986 24023 forward 0
9 Genome1 protE 24024 25010 forward 0
10 Genome1 protF 20474 22720 forward 0
11 Genome2 genA 8345 10330 forward 0
12 Genome2 genB 10327 11181 forward 0
13 Genome2 genC 11394 11843 forward 1
14 Genome2 genD 11878 12255 forward 0
15 Genome2 genE 12258 13337 forward 1
16 Genome2 genF 13365 13733 reverse 1
17 Genome2 protA 13726 14067 forward 1
(03) 基因结构绘制:
library(ggplot2)
library(gggenes)
ggplot(example_genes, aes(xmin = start, xmax = end, y = molecule, fill = gene)) +
geom_gene_arrow() +
facet_wrap(~ molecule, scales = "free", ncol = 1) +
scale_fill_brewer(palette = "Set3")
ggsave("plot1.png", plot = p1, dpi = 300, units = "in")
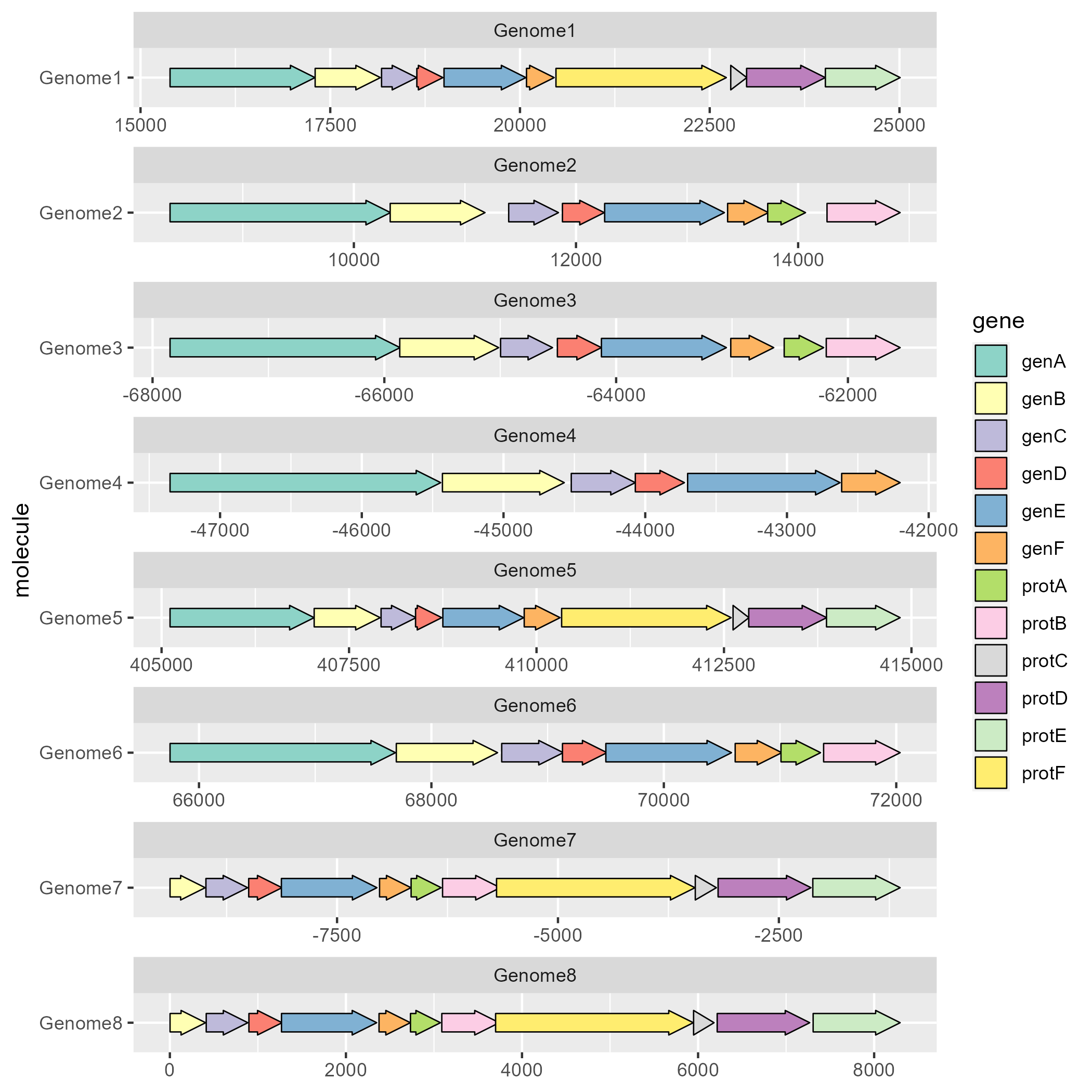
(04) 使用make_alignment_dummies()函数在各个子图中对齐基因
make_alignment_dummies()是一个辅助函数,专门用于在ggplot2的facet子图中对齐基因。在上述代码示例中,make_alignment_dummies()可以生成一个包含对齐占位符的数据框,以便在各个子图中保持基因的相对位置。这个函数在处理基因组数据时非常有用,尤其是在将基因跨多个子图可视化时需要保持相对位置一致。
dummies <- make_alignment_dummies(
example_genes,
aes(xmin = start, xmax = end, y = molecule, id = gene),
on = "genE"
)
> head(dummies)
molecule start end gene
Genome1.5 Genome1 15086 26027 genE
Genome2.15 Genome2 8345 19286 genE
Genome3.23 Genome3 -68040 -57099 genE
Genome4.31 Genome4 -47614 -36673 genE
Genome5.37 Genome5 404838 415779 genE
Genome6.47 Genome6 65588 76529 genE
p3 <- ggplot(example_genes, aes(xmin = start, xmax = end, y = molecule, fill = gene)) +
geom_gene_arrow() +
geom_blank(data = dummies) +
facet_wrap(~ molecule, scales = "free", ncol = 1) +
theme_genes()
ggsave("plot3.png", plot = p3, dpi = 300, units = "in")
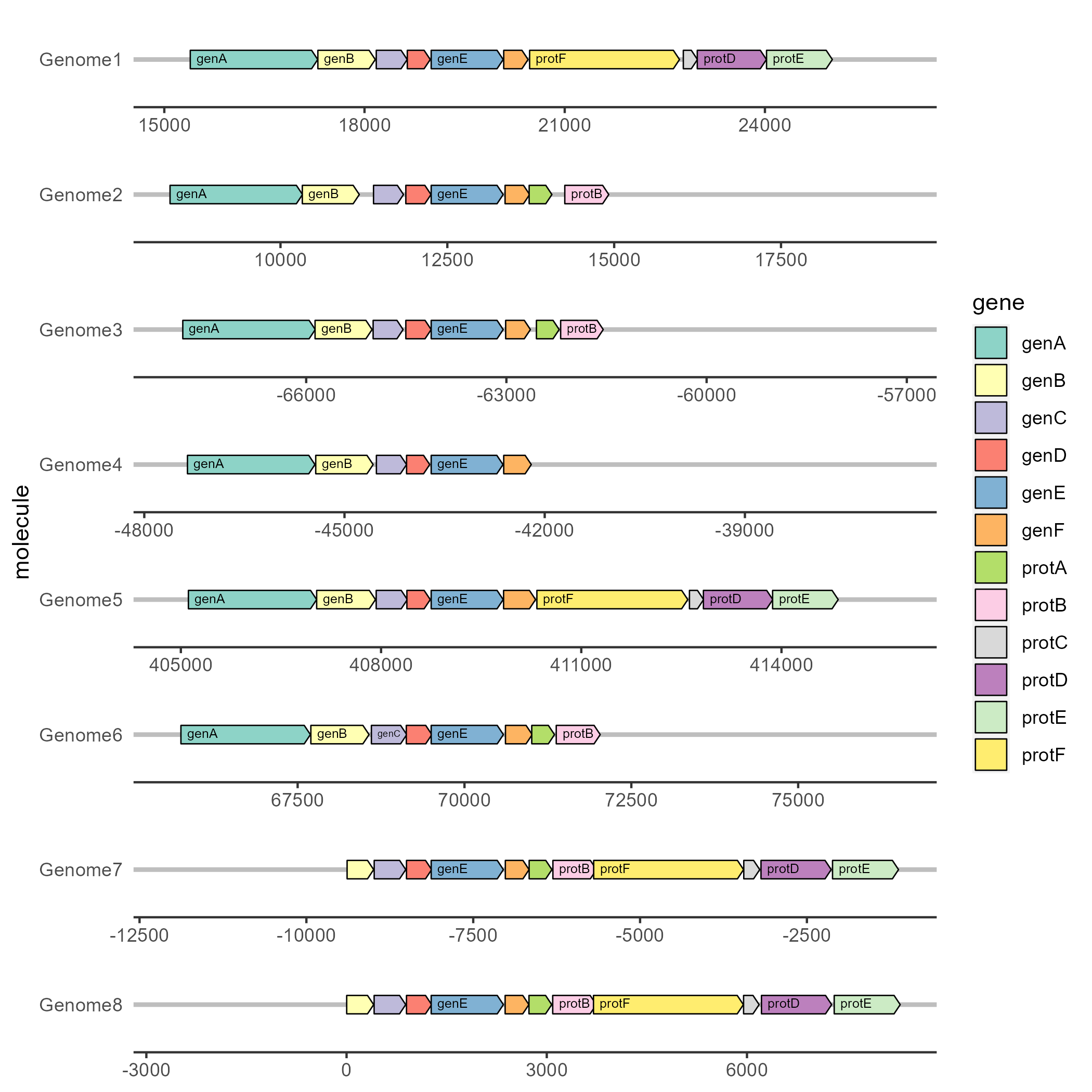
(05) 使用geom_gene_label()函数为基因添加标签
ggplot(example_genes, aes(xmin = start, xmax = end, y =
molecule, fill = gene, label = gene)) +
geom_gene_arrow(arrowhead_height = unit(3, "mm"), arrowhead_width = unit(1, "mm")) +
geom_gene_label(align = "left") +
geom_blank(data = dummies) +
facet_wrap(~ molecule, scales = "free", ncol = 1) +
scale_fill_brewer(palette = "Set3") +
theme_genes()
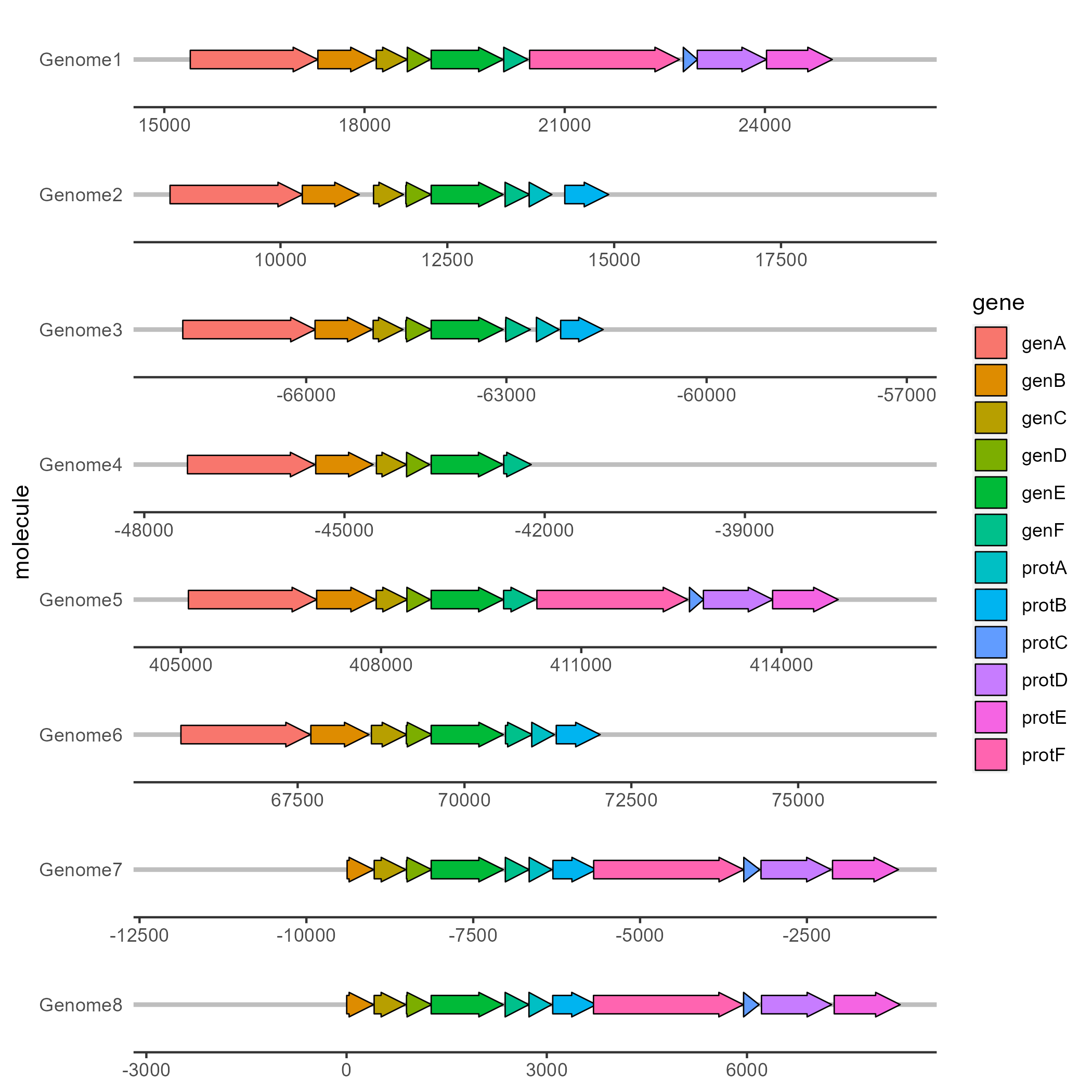
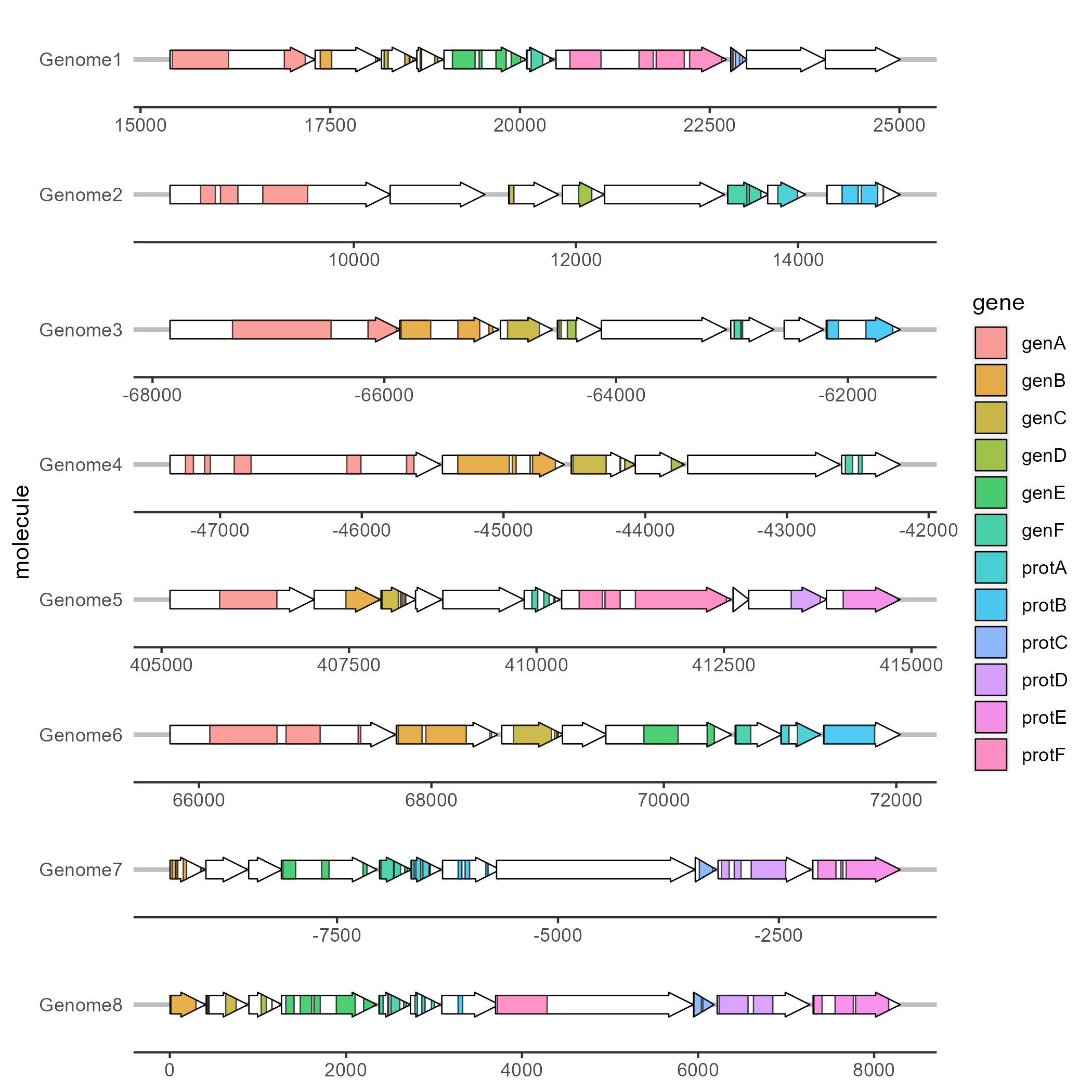
(06) 使用geom_subgene_arrow()函数展示基因内部关键区域:
可以用来展示高相似性区域、关键基因结构域或者基因组结构变异区域。
head(example_subgenes)
molecule gene start end strand subgene from to orientation
1 Genome5 genA 405113 407035 forward genA-1 405774 406538 0
2 Genome5 genB 407035 407916 forward genB-1 407458 407897 0
3 Genome5 genC 407927 408394 forward genC-1 407942 408158 0
4 Genome5 genC 407927 408394 forward genC-2 408186 408209 0
5 Genome5 genC 407927 408394 forward genC-3 408233 408257 0
6 Genome5 genF 409836 410315 forward genF-1 409938 410016 0
p5 <- ggplot(example_genes, aes(xmin = start, xmax = end, y = molecule)) +
facet_wrap(~ molecule, scales = "free", ncol = 1) +
geom_gene_arrow(fill = "white") +
geom_subgene_arrow(data = example_subgenes,
aes(xmin = start, xmax = end, y = molecule, fill = gene,
xsubmin = from, xsubmax = to), color="black", alpha=.7) +
theme_genes()
ggsave("plot5.png", plot = p5, dpi = 300, units = "in")
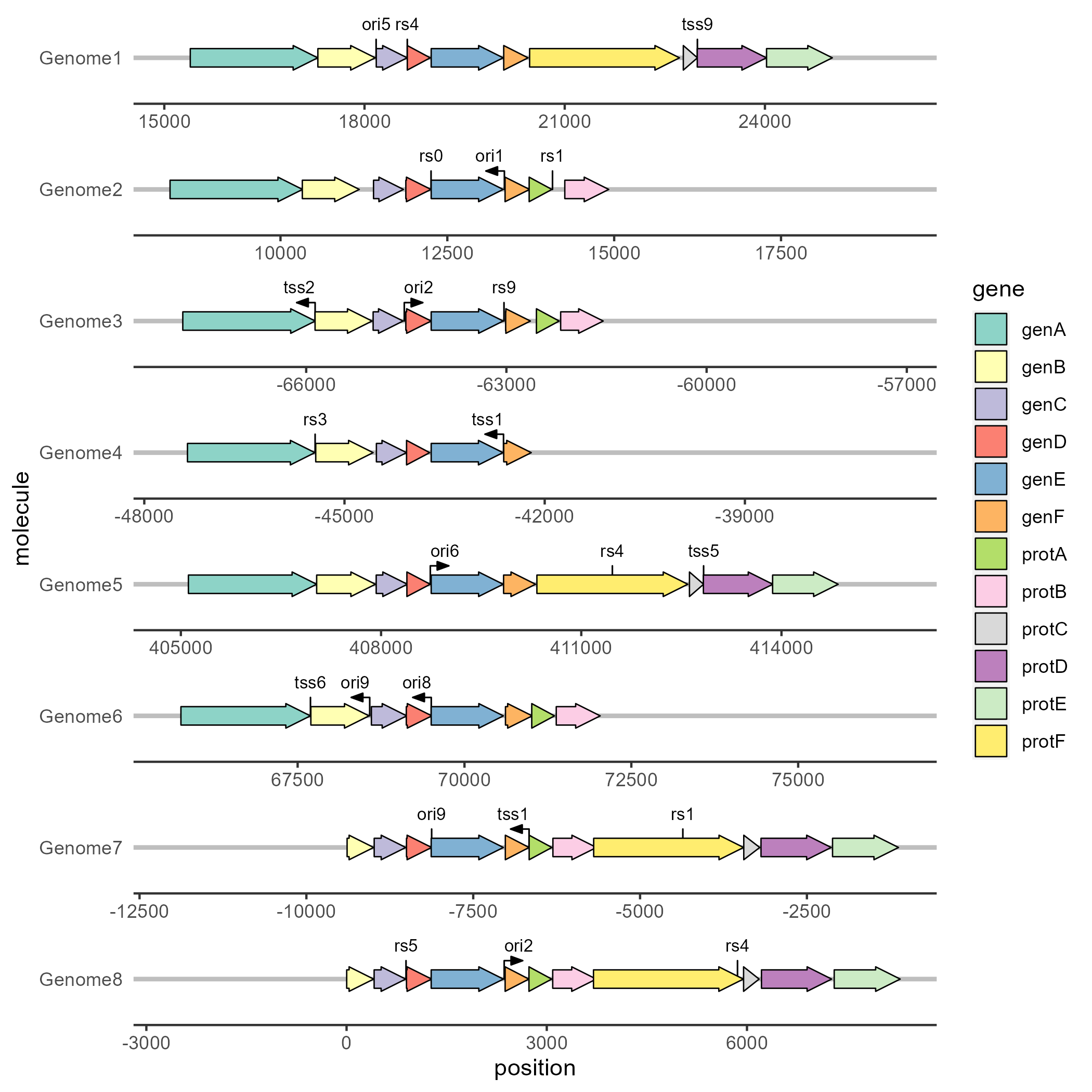
(07) 使用geom_feature()函数绘制点状基因特征:
我们可以使用geom_feature()函数绘制点状基因特征,如限制性酶切位点或转录起始位点。
> head(example_features)
molecule name type position forward
1 Genome1 tss9 tss 22988 NA
2 Genome1 rs4 restriction site 18641 NA
3 Genome1 ori5 ori 18174 NA
4 Genome2 rs0 restriction site 12256 NA
5 Genome2 rs1 restriction site 14076 NA
6 Genome2 ori1 ori 13355 FALSE
> head(example_dummies)
molecule start end gene
1 Genome1 15086 26027 genE
2 Genome2 8345 19286 genE
3 Genome3 -68040 -57099 genE
4 Genome4 -47614 -36673 genE
5 Genome5 404838 415779 genE
6 Genome6 65588 76529 genE
p6 <- ggplot(example_genes, aes(xmin = start, xmax = end, y = molecule, fill = gene)) +
geom_feature(
data = example_features,
aes(x = position, y = molecule, forward = forward)
) +
geom_feature_label(
data = example_features,
aes(x = position, y = molecule, label = name, forward = forward)
) +
geom_gene_arrow() +
geom_blank(data = example_dummies) +
facet_wrap(~ molecule, scales = "free", ncol = 1) +
scale_fill_brewer(palette = "Set3") +
theme_genes()
ggsave("plot6.png", plot = p6, dpi = 300, units = "in")